Modulation of endothelial glycocalyx and microcirculation during anaerobic exercise by supplementation with pomegranate extract

Sign up for access to the world's latest research
Abstract
Introduction: The purpose of the study was to determine whether the preferential localization of the infection and age affect the prognostic value of the genetic marker AQP5 (1364A/C, rs3759129) in outcome prediction in sepsis patients. Studies by Adamzik and colleagues have demonstrated that aquaporin AQP5 polymorphism (1364A/C, rs3759129) associates with increased 30-day survival in sepsis patients presumably due to increased gene expression that enhance the leukocyte migration. To increase the informative value of the prediction and decrease the cost, it might be crucial to determine at a pre-test level the subset of patients who might benefit most from the prognostic genotyping. Methods: Sepsis and septic shock were defined in patients according to SEPSIS-3 (2016) recommendations. Study groups (n=152) included ICU patients with abdominal sepsis (AS, including pancreatitits, peritonitis, cholecystitis, appendicitis; n=98) and sepsis patients with other sources of infections. AQP5 polymorphism was studied by analyzing PCR products in a 2% agarose gel using a AQP5 1364A/C specific tetra primer set. Data were analyzed by Kaplan-Meyer plot and Fisher test, and odds ratios were calculated. Results: Distribution of alleles (A and C) and genotypes (AA, CA and CC) AQP5 1364A/C in patients with sepsis or sepsis subgroups (sepsis with no septic shock and sepsis shock patients) versus control group (healthy volunteers) did not differ. Although there was a trend to preferential survival of sepsis patients with genotype C AQP5 despite the source of infection, only patients with AQP5 CC or CA genotype and abdominal sepsis (Sepsis-3), or a subgroup of the same AQP5 genotype experiencing septic shock, demonstrated increased 30-day survival versus AA homozygotic patients (P<0.002). Conclusions: The informative value of detecting the AQP5 CC or CA genotype for prognosis of 30-day survival versus AA homozygotic patients is increased only in abdominal sepsis patients.
Figures (379)











![Table 1 (abstract P014). Multivariate analysis for evaluating the risk of organ failure based on the LCN2 expression levels. Adjusting variables were [age], [chronic cardiac disease], [cancer], [immunosuppression], [hypertension], [chronic respiratory disease], [chronic renal failure], [respiratory focus], [abdomen focus]](https://figures.academia-assets.com/110459643/table_005.jpg)

![Table 2 (abstract P014). Multivariate analysis for evaluating the risk of mortality based on the LCN2 expression levels. Adjusting variables were [age], [chronic renal failure], [diabetes], [respiratory focus]](https://figures.academia-assets.com/110459643/figure_009.jpg)
















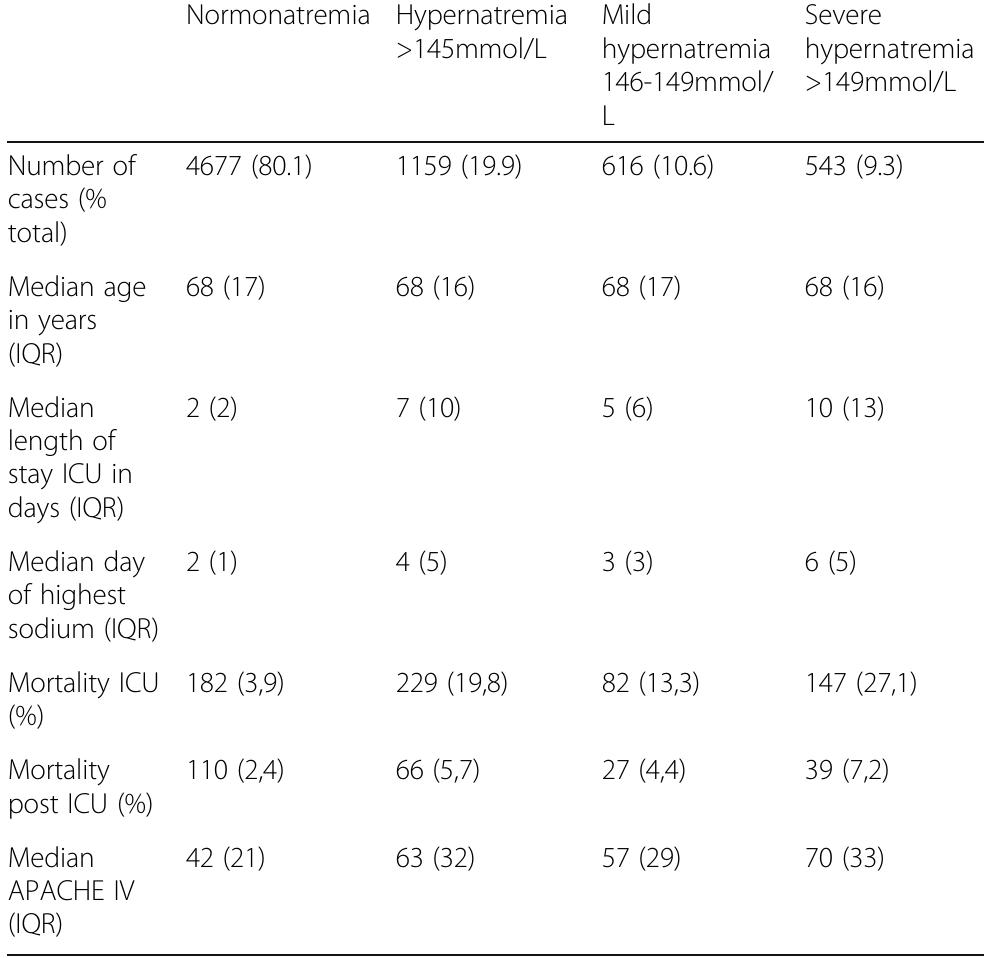
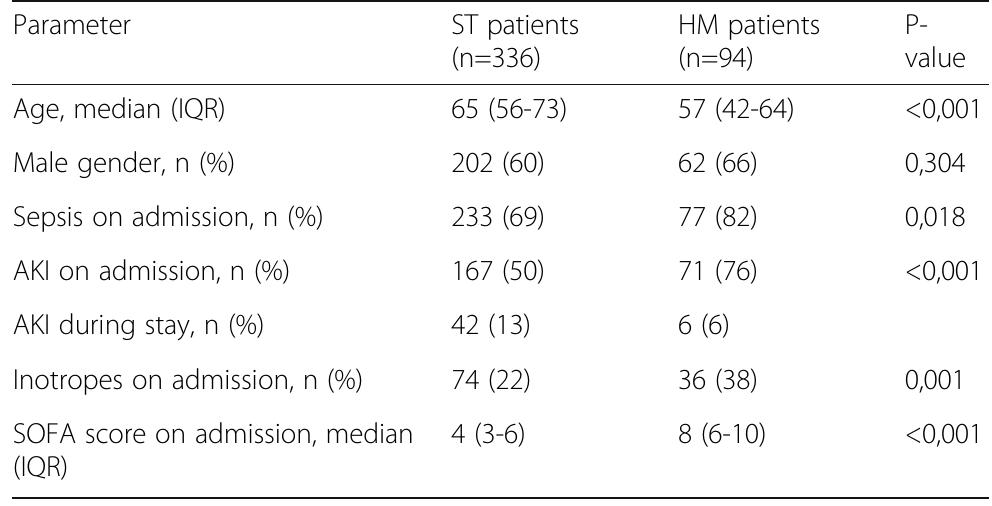
![Table 1 (abstract P036). See text for description Data presented as number (percentage) and median [interquartile range] Table 2 (abstract P036). See text for description](https://figures.academia-assets.com/110459643/table_011.jpg)



![Data presented as number (percentage) and median [interquartile range]](https://figures.academia-assets.com/110459643/figure_023.jpg)






















































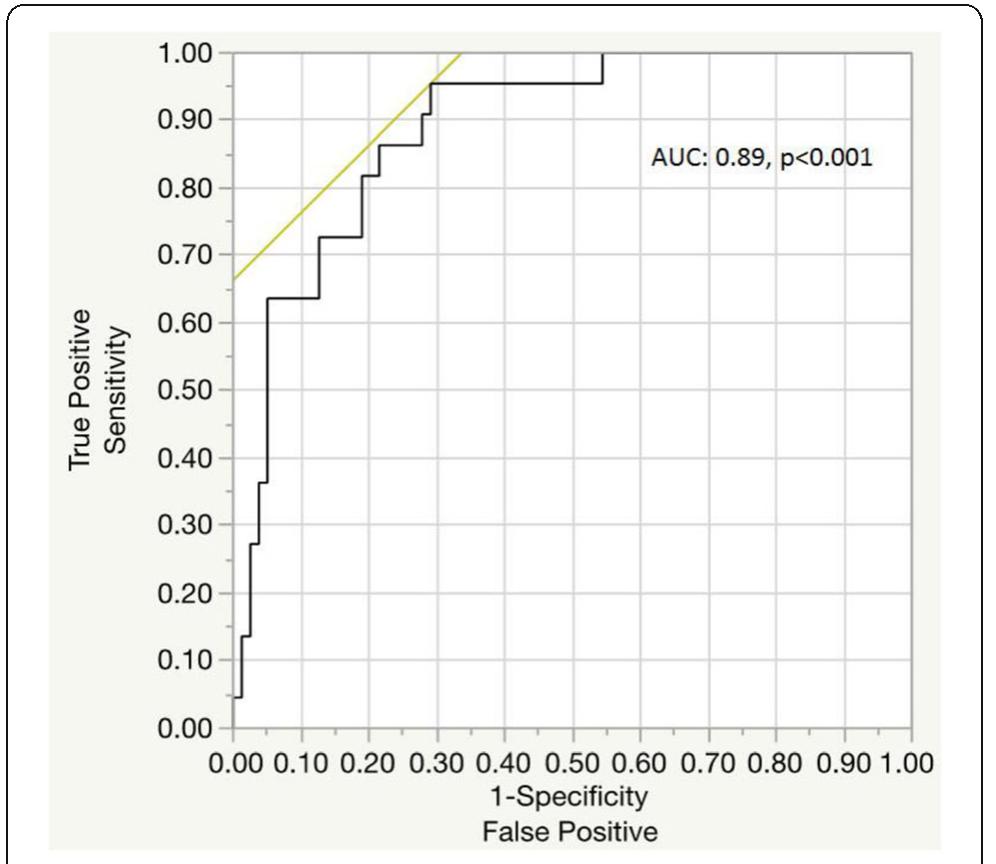






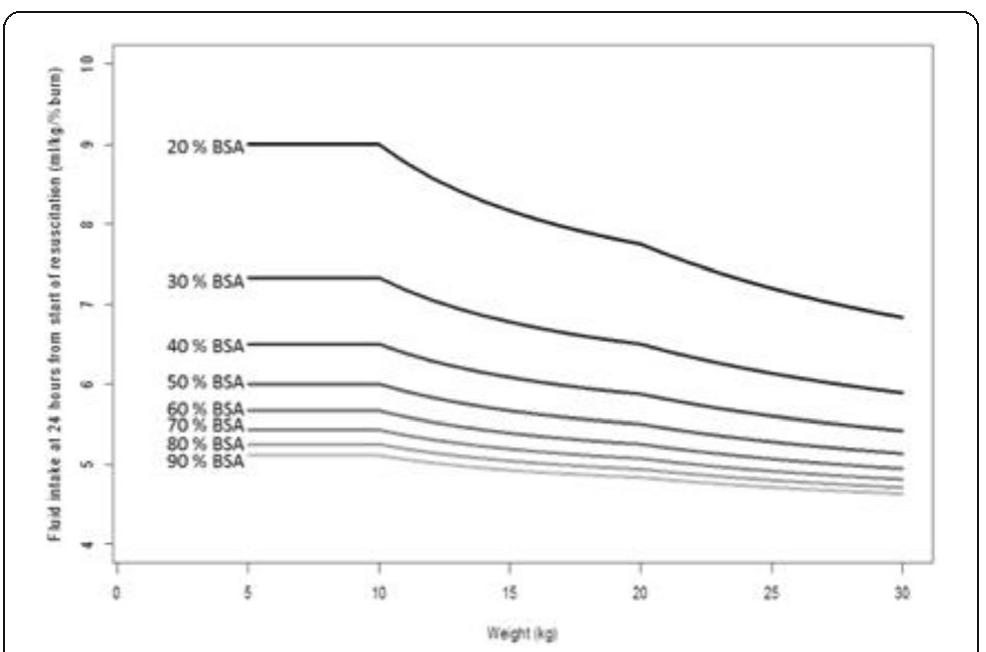










































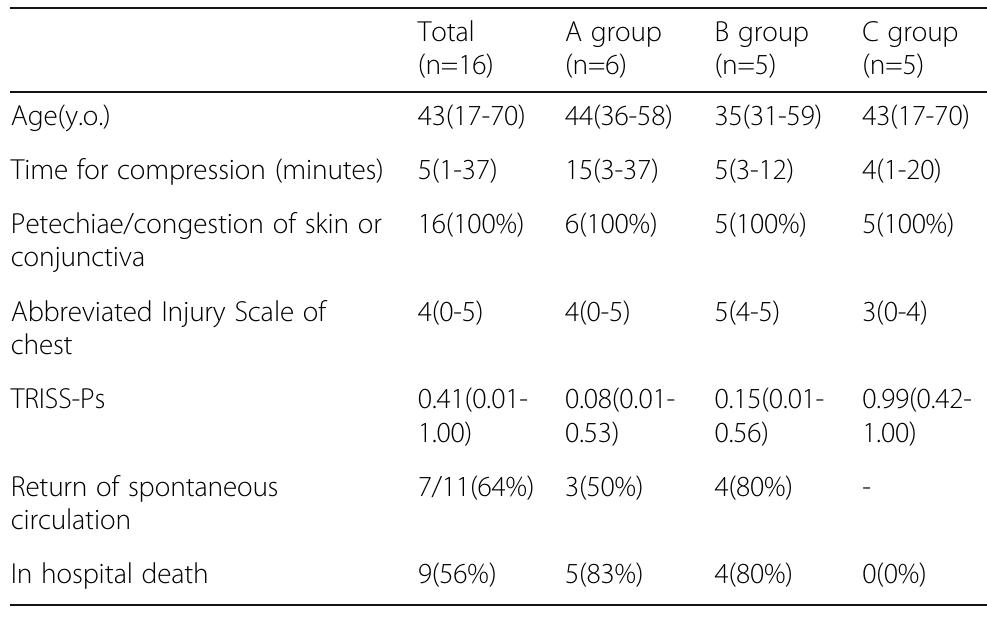

















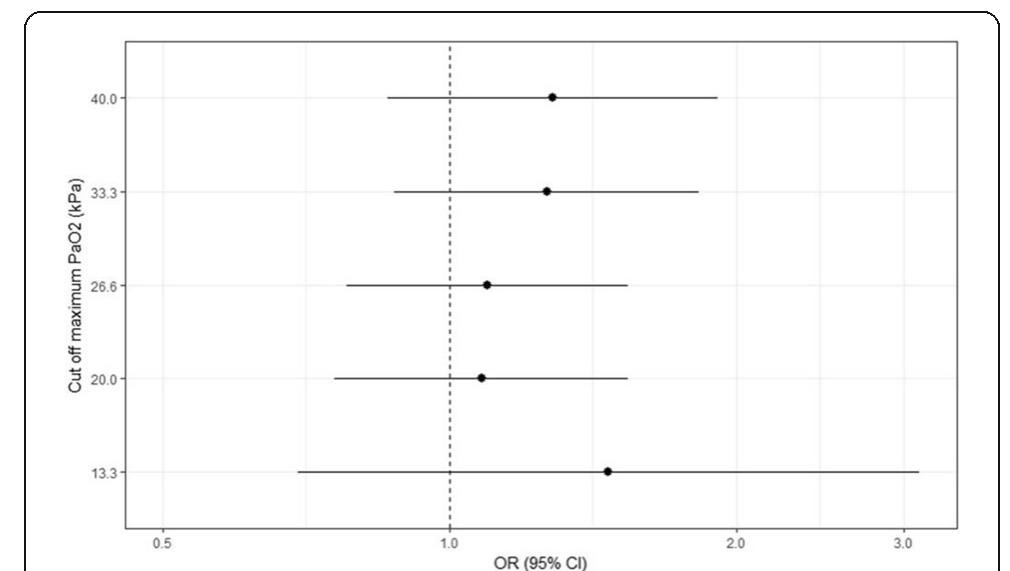
![Fig. 1 (abstract P204). Median (inter-quartile range) regional cerebral oxygen saturation (rSO2) in patients with good (Cerebral Performance Category [CPC] 1-2) and poor (CPC 3-5) neurological outcome during the first 36 h after intensive care unit (ICU) admission](https://figures.academia-assets.com/110459643/figure_099.jpg)
![Fig. 1 (abstract P205). Scatter plots of serum neuron-specific enolase (NSE) concentration at 48 h after cardiac arrest vs. median regional cerebral oxygen saturation (rSO2) during the first 36 h in intensive care unit in patients with good (Cerebral Performance Category [CPC] 1-2) and poor (CPC 3-5) neurological outcome Conclusions: We did not find any association between cerebral oxy- genation during the first 36 h of post-resuscitation intensive care and NSE serum concentrations at 48 h after cardiac arrest. Results: We did not find significant correlation between median rSOz and serum NSE concentration at 48 h after cardiac arrest, rs = -0.08, p = 0.392 (Figure 1). The median (IQR) NSE concentration at 48 h was 17.5 (13.4-25.0) ug/l and 35.2 (22.6-95.8) g/l in patients with good and poor outcome, respectively, p < 0.001.](https://figures.academia-assets.com/110459643/figure_100.jpg)
























![{1] Czosnyka M et al. J. Neurosurg., 88:802-808, 1998 Reference](https://figures.academia-assets.com/110459643/figure_115.jpg)








































![Fig. 1 (abstract P2839). Serial creatinine results pre-and post ECMO [1] Antonucci E et al. Artificial Organs 40:746-54, 2016. {2] Forni LG et al. Intensive Care Med 43:855-866, 2017. P290B P290A References](https://figures.academia-assets.com/110459643/figure_140.jpg)


















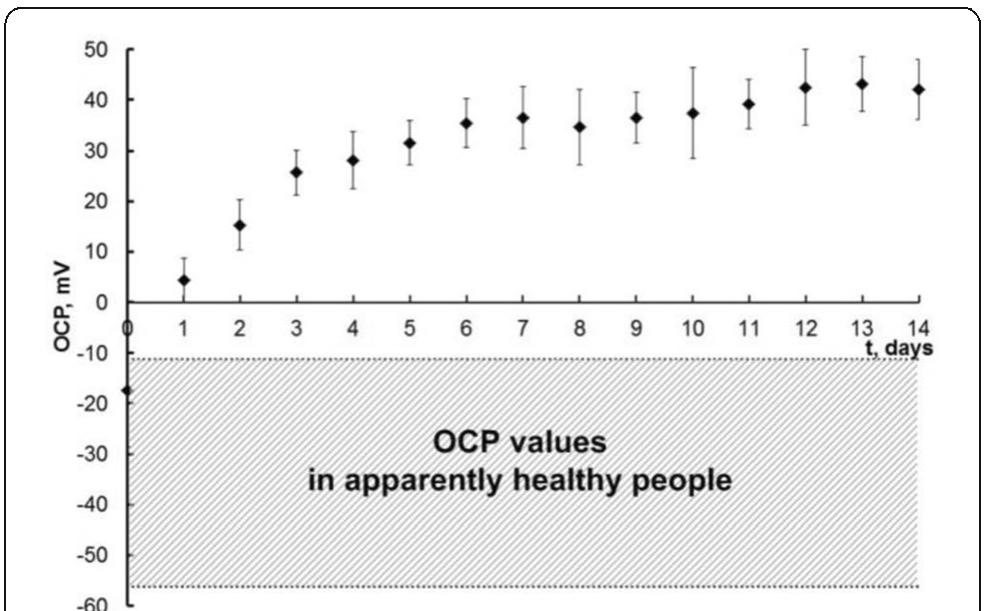



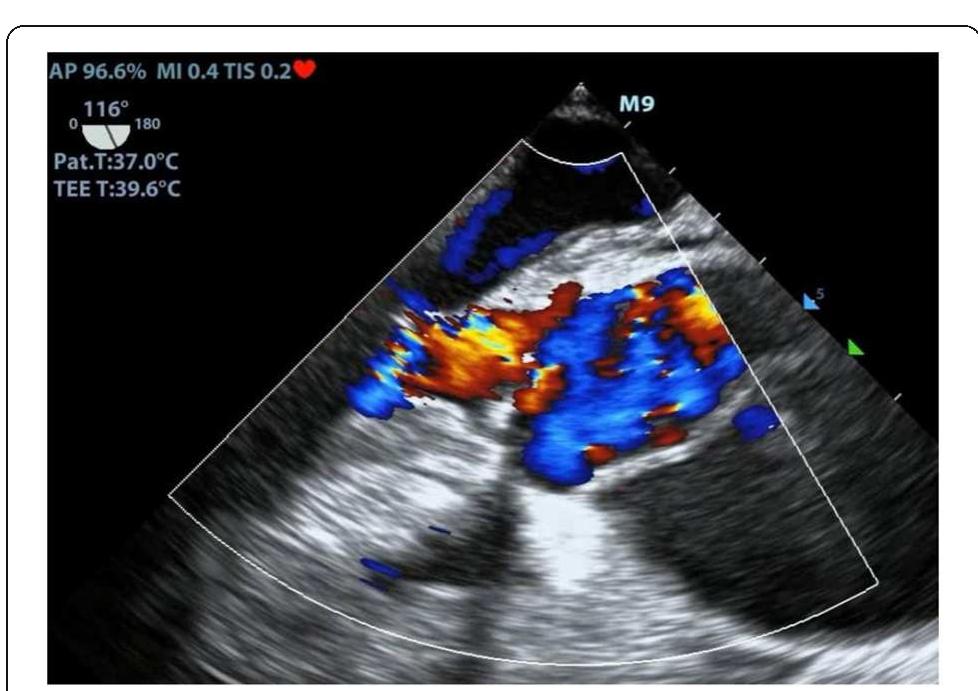




















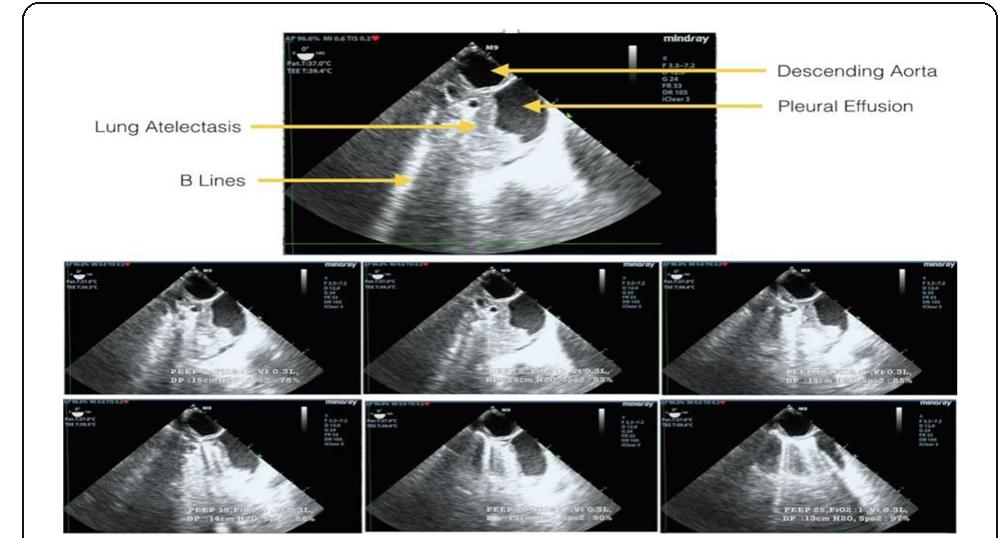
![[1] Reminiac et al. JAMPDD. 2016; 29(2):134-41. Reference](https://figures.academia-assets.com/110459643/figure_174.jpg)































































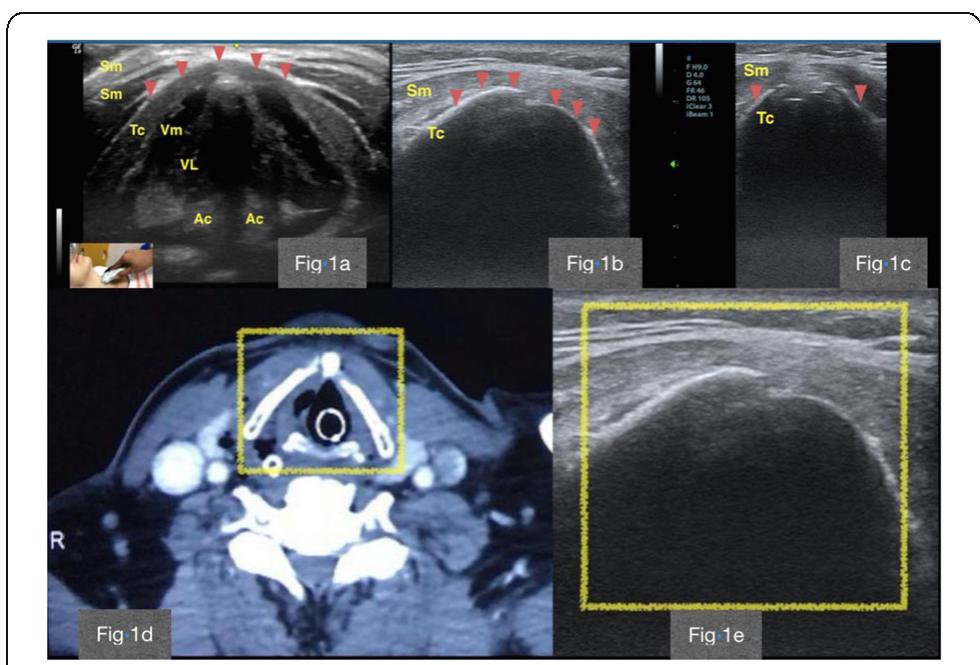
![[1] Hudson D et al. Int J Med Informatics 112, 131-136, 2018 Reference](https://figures.academia-assets.com/110459643/figure_215.jpg)









![Fig. 1 (abstract P445). Hierarchy of intervention effectiveness [from 1]](https://figures.academia-assets.com/110459643/figure_224.jpg)























Related papers
The Lancet Respiratory Medicine, 2017
der Poll, on behalf of the MARS consortium* Background Host responses during sepsis are highly heterogeneous, which hampers the identification of patients at high risk of mortality and their selection for targeted therapies. In this study, we aimed to identify biologically relevant molecular endotypes in patients with sepsis. Methods This was a prospective observational cohort study that included consecutive patients admitted for sepsis to two intensive care units (ICUs) in the Netherlands between Jan 1, 2011, and July 20, 2012 (discovery and first validation cohorts) and patients admitted with sepsis due to community-acquired pneumonia to 29 ICUs in the UK (second validation cohort). We generated genome-wide blood gene expression profiles from admission samples and analysed them by unsupervised consensus clustering and machine learning. The primary objective of this study was to establish endotypes for patients with sepsis, and assess the association of these endotypes with clinical traits and survival outcomes. We also established candidate biomarkers for the endotypes to allow identification of patient endotypes in clinical practice. The discovery cohort had 306 patients, the first validation cohort had 216, and the second validation cohort had 265 patients. Four molecular endotypes for sepsis, designated Mars1-4, were identified in the discovery cohort, and were associated with 28-day mortality (log-rank p=0•022). In the discovery cohort, the worst outcome was found for patients classified as having a Mars1 endotype, and at 28 days, 35 (39%) of 90 people with a Mars1 endotype had died (hazard ratio [HR] vs all other endotypes 1•86 [95% CI 1•21-2•86]; p=0•0045), compared with 23 (22%) of 105 people with a Mars2 endotype (HR 0•64 [0•40-1•04]; p=0•061), 16 (23%) of 71 people with a Mars3 endotype (HR 0•71 [0•41-1•22]; p=0•19), and 13 (33%) of 40 patients with a Mars4 endotype (HR 1•13 [0•63-2•04]; p=0•69). Analysis of the net reclassification improvement using a combined clinical and endotype model significantly improved risk prediction to 0•33 (0•09-0•58; p=0•008). A 140-gene expression signature reliably stratified patients with sepsis to the four endotypes in both the first and second validation cohorts. Only Mars1 was consistently significantly associated with 28-day mortality across the cohorts. To facilitate possible clinical use, a biomarker was derived for each endotype; BPGM and TAP2 reliably identified patients with a Mars1 endotype. Interpretation This study provides a method for the molecular classification of patients with sepsis to four different endotypes upon ICU admission. Detection of sepsis endotypes might assist in providing personalised patient management and in selection for trials.
AACN Advanced Critical Care, 2006
Egyptian Journal of Medical Human Genetics– Elsevier. 2010;11:97–103.
Sepsis and its sequelae are still a major cause of morbidity and mortality in intensive care units (ICU). The evidence that endogenous mediators actually mediate the individual’s response to infection has led to various approaches to assess the individual’s contribution to the course of the disease. The role of an individual’s genetic background and predisposition for the extent of inflammatory responses is determined by variability of genes encoding endogenous mediators that constitute the pathways of inflammation. Pro- and anti-inflammatory responses contribute to the susceptibility and outcome of patients with systemic inflammation and sepsis. Therefore, all genes encoding proteins involved in the transduction of inflammatory processes are candidate genes to determine the human genetic background that is responsible for interindividual differences in systemic inflammatory responses. Polymorphism of TNA a, TNFB, IL-10, IL-6, IL-1b, IL-1RN, HMGB1, TLR, PAI-1, DEFB1, HSP and MMP-9 has contribution to susceptibility and outcome of sepsis in some research. Examination of the association between genetic polymorphisms and sepsis promises to provide clinicians with new tools to evaluate prognosis, to intervene early and aggressively in treating high risk persons, and to avoid the use of therapies with adverse effects in treating low risk persons. Genomic information may be useful to use to identify groups of patients with a high risk of developing severe sepsis and multiple organ dysfunctions.
EBioMedicine, 2016
Sepsis is the dysregulated host response to an infection which leads to life-threatening organ dysfunction that varies by host genomic factors. We conducted a genome-wide association study (GWAS) in 740 adult septic patients and focused on 28 day mortality as outcome. Variants with suggestive evidence for an association (p ≤ 10 −5) were validated in two additional GWA studies (n = 3470) and gene coding regions related to the variants were assessed in an independent exome sequencing study (n = 74). In the discovery GWAS, we identified 243 autosomal variants which clustered in 14 loci (p ≤ 10 −5). The best association signal (rs117983287; p = 8.16 × 10 −8) was observed for a missense variant located at chromosome 9q21.2 in the VPS13A gene. VPS13A was further supported by additional GWAS (p = 0.03) and sequencing data (p = 0.04). Furthermore, CRISPLD2 (p = 5.99 × 10 −6) and a region on chromosome 13q21.33 (p = 3.34 × 10 −7) were supported by both our data and external biological evidence. We found 14 loci with suggestive evidence for an association with 28 day mortality and found supportive, converging evidence for three of them in independent data sets. Elucidating the underlying biological mechanisms of VPS13A, CRISPLD2, and the chromosome 13 locus should be a focus of future research activities.
The Journal of Infection in Developing Countries, 2012
Introduction: A patient's response to sepsis is influenced by their genetic background. Our objective was to use plasma markers, such as protein C (PC), D-dimer, Plasminogen Activator Inhibitor-1 (PAI-1) levels, and the PAI-1 rs1799889 4G/5G and Tumor Necrosis Factor-α rs1800629 G/A polymorphisms to improve classical intensive care unit (ICU) scores. Methodology: We studied 380 subjects, 166 with sepsis. We performed coagulation tests: plasma PAI-1 and PC levels were evaluated by chromogenic methods; and D-dimer was evaluated by immunoturbidimetric assay. Polymorphisms were performed using for polymerase chain reactions followed by digest with specific restriction enzyme. We acquired the APACHE and SOFA scores (time zero), sex, age, body mass index, associated co-morbidities, length of ICU stay (days), the severity of sepsis (sepsis, severe sepsis or septic shock), the HIV status and the ICU outcome (survival or death). Results: We found significant differences between patients who died (n=80) and those who survived (n=86) in terms of the ICU length of stay (6 vs. 10 days), septic shock (64 versus 24%), age (51 versus 38 years old), HIV+ condition (34 versus 16%), SOFA (7 versus 4), APACHE (19 versus 13), D-dimer (4.32 versus 2.88 g/ml), PC (46.0 versus 63.5 %) and PAI-1 (33.0 versus 16.5 UA/l). When we used a regression analysis with dichotomized variables, only the SOFA 4 , PAI-1 16 , HIV status and the PAI-1 4G allele proved to be predictors of death at time zero. Conclusions: In the future, ICU scores may be further improved by adding certain genomic or plasma data.
2018
The purpose of this study was to determine the impact of baseline systemic inflammation (pro-inflammatory cytokine, anti-inflammatory cytokine, and their ratio), genetic variability, and environment on the development of health care associated infections (HAI) among sepsis patients during their ICU stay (up to 28 days). Methods: A prospective observation study was conducted at the Veterans Affairs Medical Center in the Medical Intensive Care Unit over an 18 month period. A total of 78 patients were enrolled within 72 hours of presenting to the ICU with sepsis. Patient were excluded if they were receiving immunosuppressants (chemotherapy or greater than one mg/kg of prednisone or equivalent dose), immunosuppressed (AIDS, cancer), or had liver failure (Child Pugh category C or higher). Baseline plasma and buccal swabs were collected. Patients were followed prospectively through their ICU stay (or for a maximum of 28 days) for the development of HAI as defined by CDC guidelines. Primary variables included baseline IL-6 and IL-10 levels, IL-6 SNP rs1800795, IL-10 SNP rs1800896, APACHE II, invasive devices, and development of HAI. Results: A total of 17 HAI were identified with 64% caused by Candida. There were no significant differences in levels of pro-inflammatory cytokine, anti-inflammatory cytokine, or their ratio among subjects who did and did not develop at least one HAI during their ICU stay. There were also no significant differences in rs1800795 or rs1800896 genotypes for those who did and did not develop HAI; however, racial differences were detected in genotypes among white and black patients with sepsis who did and did not develop HAI. There was a significant difference in rs1800795 genotype among black patients with sepsis who did not develop HAI compared to whites patients with sepsis who did not develop HAI (p = 0.006). Specifically, black patients had a lower CG (17.4% vs. 42.1%) and higher GG (82.6% vs. 42.1%) than white patients. There were no racial differences when comparing white and black sepsis patients who developed HAI (p = 1.0). In a series of Cox regression analyses investigating timing to first HAI among those who did and did not develop HAI during ICU stay, the final model included only APACHE II, cumulative invasive device score, and IL-6 rs1800795. Conclusion: This study provides evidence of a genetic risk for development of HAI. Despite best evidenced based practices some patients will develop HAI. Strict aseptic technique is essential to preventing infection. In addition to eliminating invasive devices as quickly as possible, patients with a high severity of illness may need to be isolated to lower their risk. Early administration of antibiotics not only provides prompt treatment for the initial infection but also lowers risk for subsequent infections. vii TABLE OF CONTENTS CHAPTER 1.
Critical Care, 2004
Sepsis is a complex syndrome that develops when the initial, appropriate host response to an infection becomes amplified, and is then dysregulated. Among other factors, the innate immune system is of central importance to the early containment of infection. Death from infection ...
Indian Journal of Critical Care Medicine, 2019
Objective: The outcomes of sepsis and septic shock patients are heterogonous, with a variable response despite standardized care. The aim of this study was to explore the racial differences in septic shock outcomes, and their association with genetic polymorphisms and cytokine levels in an Asian population. Design: This was an observational cohort study, Setting: Intensive Care units of a 500 bedded tertiary care hospital in Singapore. Patients: 198 patients (73 Chinese, 73 Malay and 52 Indian and others) admitted to the Khoo Teck Puat Hospital Intensive Care Unit between August 2016 and June 2017, with a diagnosis of severe sepsis (according to) were enrolled. Interventions: Plasma interlukin-6 (IL-6), interlukin-10 (IL-10) and Tumour necrosis factor-a (TNFa) were measured using a highly sensitive quantitative sandwich enzyme-linked immunosorbent assay (ELISA) (BioVendor, Modrice, Czech Republic). The gene panel studied included 16 genes. Measurements and Main Results: The rs7038903 common variant in SVEP1 gene showed significant association with sepsis severity independent of other variants in ordinal logistic and linear regression model (p=0.001 and p=0.002 respectively). Moreover, the association between rs7038903 and increased hazard for death remained significant after further adjusting for cytokines level. Interestingly, significant differences were seen in plasma IL6 in individuals with or without rs7038903 C allele (28pg/ml (IQR 12-86) vs 90pg/ml (IQR 49-155); P=0.022) in patients with severe sepsis in the Malay ethnic group. Conclusions: Our study shows a promising polymorphism in SVEP1 gene (rs7038903) which is associated with sepsis shock and 28 days mortality, independent of age, gender, and method of diagnosis and SOFA score. Collectively, while our findings so far have shown the additional value or measuring cytokines and genetic markers in sepsis outcomes in the local population, further large scare studies are needed in a heterogeneous septic population with a rigorous analysis to know the significance of our findings.
Clinical Infectious Diseases, 2005
Genetic variation has been shown to play a large role in determining susceptibility to and outcome of such complex diseases as sepsis. There is a much higher heritability of death due to infection than death due to cancer or heart disease. More than 8 million single nucleotide polymorphisms (SNPs) have been detected in the human genome, and there is very little understanding of their effect on gene expression and protein function. The use of haplotypes, which are inherited sets of linked SNPs, as the unit of genetic variation in association studies and the marking of these haplotypes with unique "tag SNPs" may help to narrow down the search for causal SNPs. Future studies must be large (thousands of patients) and must be carefully designed to avoid false associations resulting from ethnic differences in genotype frequencies and disease prevalence in order to find true, reproducible associations between genotype and phenotype. Functional studies and careful characterization of intermediate phenotypes must be done to lend biological plausibility to genotype-phenotype associations. Examination of the association between genetic polymorphisms and sepsis promises to provide clinicians with new tools to evaluate prognosis, to intervene early and aggressively in treating high-risk persons, and to avoid the use of therapies with adverse effects in treating low-risk persons.
Critical Care Medicine, 2008

Loading Preview
Sorry, preview is currently unavailable. You can download the paper by clicking the button above.
Related papers
Expert Review of Anti-infective Therapy, 2012
Prague medical report, 2008
Critical Care Medicine, 2003
JAMA, 2019
Biomarker Insights, 2019
eBioMedicine, 2022
Critical Care, 2014
Critical Care, 2019
Laboratory Medicine
The Lancet. Respiratory medicine, 2015
Journal of Translational Medicine, 2009
 Zivile Pranskuniene
Zivile Pranskuniene
